Oligonucleotides Guide
Table of Contents
9. Applications of Oligonucleotides
9.1. Oligonucleotides as PCR Primers
9.2. Sequencing using Oligonucleotides
9.3. Use of Oligonucleotides in Gene Expression Studies
9.4. Antisense Oligonucleotides
9.5. Oligonucleotide Therapeutics
9.6. Unraveling cellular function with oligonucleotides is a key step toward creating new drugs
9.7. World’s data can be stored in oligonucleotides for 10,000 years
9.8. Other uses of Oligonucleotides
10. What's Next?
11. References
1. Introduction
Oligonucleotides, or oligos, are among the most useful materials in contemporary molecular biology. They are arguably the single most useful instrument in contemporary molecular biology. There would be no modern biotechnology, diagnostics, or pharmaceuticals without oligos.
'Oligonucleotide' comes from the Greek word 'oligo', which means few or little. The term "mer," from the Greek for "part," is commonly used to designate the length of the oligonucleotide. Oligonucleotides are a type of short nucleic acid polymer with applications in forensics, genetic testing, and other scientific fields. The purpose of oligonucleotides is to hybridize precisely to DNA or RNA sequences, and they typically consist of 13 to 25 nucleotides. These fragments of nucleic acid are produced via solid-phase clinical synthesis and then employed in applications like as polymerase chain reaction (PCR), DNA sequencing, library creation, and synthetic gene synthesis.1
A molecule of oligonucleotides may be identified by the specific order of the nucleotide residues that compose it. The oligonucleotide length is often indicated by the prefix "-mer" (from Greek meros, "part"). As an illustration, an oligonucleotide consisting of six nucleotides (nt) is called a hexamer, whereas a 25-mer is the more common term. Sequence-specific binding of oligonucleotides to their corresponding oligonucleotides, DNA, or RNA is simple and results in the formation of duplexes or, less frequently, hybrids of a higher order. This fundamental characteristic underpins the application of oligonucleotides as probes for the detection of targeted regions of DNA and RNA. Oligonucleotides are used in several diagnostic and research methods, including as polymerase chain reaction (PCR), Southern blotting, ASO analysis, fluorescence in situ hybridization (FISH), and the creation of synthetic genes.2
A variety of pharmacological effects may be produced by modifying oligonucleotides, which are made up of 2'-deoxyribonucleotides (oligodeoxyribonucleotides), either at the backbone or at the 2' sugar position. These alterations confer novel features onto the oligonucleotides, elevating them to a pivotal role in antisense treatment.3
2. History of Oligonucleotides
The term "molecular biology" was first used in 1938 by Rockefeller Foundation researcher Warren Weaver. The phrase originally referred to the application of scientific methods from the physical sciences to the study of living things. The essential foundations of molecular biology, outlining how DNA instructions are translated into RNA and proteins, were hammered out during the subsequent decades. Watson and Crick's 1953 X-ray crystallographic resolution of the DNA double helix paved the way for synthetic nucleic acid and helped scientists better grasp how DNA replication worked.
Rapid progress has been made in understanding DNA's biological functions since its molecular structure was determined and reported in 1953. The term "oligonucleotide" was used by the scientific community in order to define these molecules after the first attempts at creating synthetic DNA strands were reported in 1955. One of the founders of the technology, Martin Caruthers, provides a lovely recounting of the complex history of DNA synthesis chemistry in his open-access publication. The chemistry of modern DNA synthesis, developed by Caruthers, has not altered much in over 30 years. The fields of molecular biology, synthetic biology, and biotechnology have all grown to rely heavily on oligos. The reason is, both manufactured DNA and naturally occurring DNA have the same chemical and structural properties. DNA synthesized through phosphoramidite chemistry is identical to DNA synthesized within a cell in every way except for its origin.
Because of its role in the polymerase chain reaction, oligonucleotides have been making waves in research for over 30 years (PCR). The PCR process is analogous to making a photocopy of a book page using DNA. In PCR, the photocopier (an enzyme called DNA polymerase) must be introduced to the paper (the DNA) in order to create copies, in contrast to the usual photocopying process.4
3. Structure of Oligonucleotides
Every oligonucleotide has three parts: a nucleotide or base (purine or pyrimidine), a pentose five-carbon sugar (ribose for RNA or deoxyribose for DNA), and a phosphate group. The nucleic acid backbone consists of the phosphate groups connecting adjacent monomers (individual nucleotides) and the sugar moieties. Each nucleic acid strand is oriented in a certain way, with the directionality flowing from the 5′-end to the 3′-end.
The nucleotides in double-stranded DNA link up with their complementary bases because of the strands' opposing orientations. Purines include adenine and guanine, whereas pyrimidines include cytosine, thymine, and uracil (all found in DNA) (U; in RNA). Adenine forms hydrogen bonds with either thymine or uracil, a phenomenon known as complementarity; guanine and cytosine, on the other hand, connect with each other. Each nucleotide can be further subdivided into mononucleotides, dinucleotides, oligonucleotides, and polynucleotides.5
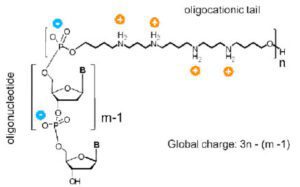
4. How long are Oligonucleotides?
The number of individual nucleotides determines an oligonucleotide's length. The term "20-mer" is used to describe a sequence of 20 nucleotides for purposes of nomenclature. When it comes to various uses, the oligo length is essential. In polymerase chain reaction (PCR), for instance, short-mer oligos are typically utilized, but very long sequences (200 bases or more) are typically reserved for demanding applications like cloning or CRISPR editing. Custom, long-mer oligos are difficult to synthesize, and it takes careful monitoring and purification to ensure a consistent sequence is delivered. In general, the length of an oligo is fewer than 60 monomers. The chance of creating undesirable by-products, which may include deletion or truncation sequences, increases with the length of the necessary sequence due to the nature of the way oligos are produced, which involves repeating reaction cycles that add extra nucleotides.4
5. How Oligonucleotides are made?
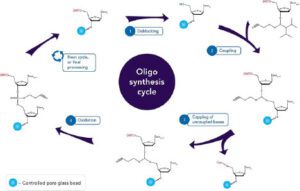
The synthesis of oligonucleotides takes place in a solid-phase chemical reactor. This involves the sequential addition of the four nucleic acids A, T, C, and G to build a developing chain of nucleotides. The chain lengthens from 3' to 5' when nucleotides are added. The finished product is an oligo of full length.6
The chemical synthesis of oligonucleotides, or segments of nucleic acids having a well-defined chemical structure, is known as oligonucleotide synthesis (sequence). The method's fast and low-cost availability of oligonucleotides with a specified sequence makes it a valuable tool in modern laboratory practise. Chemical oligonucleotide synthesis can proceed in either the 5' to 3' or 3' to 5' orientation, but enzymatic synthesis of DNA and RNA is restricted to the former. Phosphoramidite technique and phosphoramidite building blocks generated from protected 2'-deoxynucleosides (dA, dC, dG, and T), ribonucleosides (A, C, G, and U), or chemically modified nucleosides, e.g. LNA or BNA, are used in current implementation of the process as solid-phase synthesis.
Sequentially coupling the building blocks to the expanding oligonucleotide chain in the order specified by the product sequence yields the desired oligonucleotide (see Synthetic cycle below). Since the 1970s, when automation first began, the process has been completely mechanised. When the construction of the chain is complete, the product is discharged into solution, where it is deprotected and eventually collected. As the number of mistakes rises with the length of the oligonucleotide being synthesised, the occurrence of side reactions places practical restrictions on the length of synthetic oligonucleotides (up to roughly 200 nucleotide residues). High-performance liquid chromatography (HPLC) is commonly used for product isolation to create very pure oligonucleotides. Most synthetic oligonucleotides have 15-25 bases and are single-stranded RNA or DNA.7
Many fields of molecular biology and medicine make use of oligonucleotides. Primers for DNA sequencing and amplification, probes for detecting complementary DNA or RNA via molecular hybridization, tools for the targeted insertion of mutations and restriction sites, and the synthesis of artificial genes are all popular applications of oligonucleotides. Oligonucleotide synthesis is being used increasingly to recreate viruses from sequence alone, whether they are innocuous (like Phi X 174) or harmful (like the 1917 influenza virus or SARS-CoV-2).
6. Classification of Oligonucleotides
Depending on their application, oligonucleotides (oligos) may be left unmodified or treated with a variety of chemical modifications, such as the addition of 5' or 3' phosphate groups to facilitate ligation or block extension, labeling with radionuclides or fluorophores and/or quenchers for use as probes, the incorporation of thiol, amino, or other reactive moieties to facilitate the covalent coupling of functional molecules, such as Although DNA oligos have seen the more widespread application, RNA oligos are also commercially accessible. The number of oligomers is often indicated by adding the suffix -mer to the word. A 19-mer is an example of an oligonucleotide having this number of bases. The majority of applications for oligonucleotides need them to form a base pair with an existing DNA or RNA strand.8
The quantity of sugar units they contain determines their category. The list below details some of these:
- Trisaccharides: it is a kind of oligosaccharides, consisting of three monosaccharides that have been bonded together. Raffinose is a trisaccharide made up of three monosaccharides (glucose, galactose, and fructose).
- Tetrasaccharides: These have four sugars in their structure. Sesamose is a tetrasaccharide consisting of two galactose, one fructose, and one glucose.
- Pentasaccharides: These are long chains of sugars made up of five monosaccharides. One such sugar is called verbascose, which consists of three galactose units, one fructose unit, and one glucose unit.
- Hexasaccharides: There are six sugar units in hexasaccharides.
- Heptasaccharides: These consist of seven sugar units, whereas octasaccharides comprise eight sugar units, etc.9
7. Examples of Oligonucleotides
Raffinose family of oligosaccharides
A trisaccharide with the formula C18H32O16, raffinose is composed of three sugar units: fructose, glucose, and galactose. By hydrolyzing it with the -galactosidase enzyme, it yields D-galactose and sucrose. You can find it in foods including beans, beets, cabbage, cottonseed, broccoli, and more.
Fructooligosaccharides
In plants, oligosaccharides called fructooligosaccharides are produced. They are classified as storage saccharides and are also known as oligofructans. Plants typically contain these short chains of fructose residues. Agave tequilana, Helianthus tuberosus, bananas, onions, wheat, barley, etc. are all examples of such plants. In the food industry, they are used for aesthetic purposes (to change the color or texture of the dish) and for functional purposes (as artificial sweeteners).
Galactooligosaccharides
Galactose molecules are the building blocks of galactooligosaccharides. Human milk oligosaccharides are one such example (HMOs). Breast milk contains these. Lactose is the starting point for them. One-third of all HMOs are 2′-fucosyllactose, which is a fucose-galactose-glucose trisaccharide.9
8. How Oligonucleotides Function
Oligonucleotides achieve their effects by pairing up with sequences that are complementary to themselves. Oligos have various applications, but PCR, microarray analysis, in situ hybridization, and biosensing are some of the best approaches to analyzing gene expression and gene regulation. Primer strand oligonucleotides are a common application of oligonucleotides. As DNA polymerases can only add nucleotides to an already existing nucleic acid sequence, primers, which are single-stranded oligos, are necessary for the commencement of DNA replication in PCR. In polymerase chain reaction (PCR), DNA polymerase joins to the target sequence that has been annealed by the primers. Because DNA is a double helix, most PCR methods call for a pair of primers—one forward and one reverse—to generate a complementary strand.
Primers for DNA sequencing are often oligonucleotides. Traditional sequencing methods, including polymerase chain reaction (PCR), rely on DNA polymerases that need template DNA to begin elongation. Hence, primers for sequencing are DNA oligos designed to anneal to a certain target sequence. In addition to their use in recombinant DNA construction and as molecular probes in various diagnostic procedures (such as DNA microarrays, Southern blotting, an antisense oligonucleotide (AS) analysis, and fluorescent in situ hybridization), oligonucleotides are also a valuable tool in the field of molecular biology (FISH).
These methods are only possible because oligonucleotides possess unique characteristics that allow them to be used in certain ways. Particularly important to oligos' utility is their complementarity. During replication and transcription, the nucleic acid sequence is protected by a phenomenon known as complementarity, which is defined as the chemical recognition and particular hydrogen bonding between nucleotide bases. In procedures requiring the hybridization of target molecules, complementary base pairs are useful because they determine sequence specificity and preserve sequence integrity by locating each other during the annealing, or hybridization, process to generate double-stranded nucleic acid molecules.10
9. Applications of Oligonucleotides
Oligonucleotides, or oligos, are useful in a wide variety of contexts, including basic science, illness diagnostics, and, more recently, therapy. Now that cheap synthetic single-stranded DNA and RNA are widely available, DNA amplification by PCR may be performed routinely. The research of gene expression and disease processes has been greatly advanced by the use of oligonucleotides as primers. Primer-based technologies are rapidly expanding and may be used to characterize, track, and measure almost everything related to nucleic acids. Oligonucleotides have several applications outside of their primary use as polymerase chain reaction (PCR) primers. They may be employed as probes, in microarrays, in situ hybridization, in antisense analysis, and even as drug carriers.11
Following are some important applications of Oligonucleotides:
- PCR Primers
- Real-time PCR with fluorescently labeled Oligonucleotides
- RNA, siRNA and antisense studies
- FRET
- Melting point optimization of oligonucleotides (e.g. using BNAs)
- DNA/DNase work
- TLR (Toll-like receptors) research
- Multiply-primed rolling circle amplification
- Expression of Gene and Gene Therapy
- Molecular diagnostics
- Protein- protein or protein – DNA binding Interaction studies
- Inhibition of targeted protein function
- Microarrays
- Sequencing
- Antisense Oligonucleotides
- Fluorescence in Situ Hybridization (FISH)12
9.1. Oligonucleotides as PCR Primers
Short synthetic oligonucleotides (up to 30 meters in length) are used most frequently as probes and primers in a wide range of scientific disciplines. To do this, a nucleotide sequence that is complementary to a bigger, desired DNA or RNA strand must be synthesized (target sequence). Oligos are often employed as primers to kick off enzymatic processes, such as the production of millions or billions of copies of a short or lengthy target sequence. Examples include the widely used polymerase chain reaction (PCR) and the Sanger sequencing technique. DNA sequencing, gene expression, cloning, and molecular diagnostics are just a few examples of where oligo primers are put to use.
Oligos are used as probes to detect and verify the presence of a certain target sequence in a sample of DNA or RNA. The use of oligo probes is widespread, and some examples include northern blotting (for RNA) and southern blotting (for DNA), the use of fluorophore-conjugated sequences in microarrays to detect changes in gene expression, and the use of oligo probes in screening for genetic diseases or identifying specific pathogens (molecular diagnostics).13
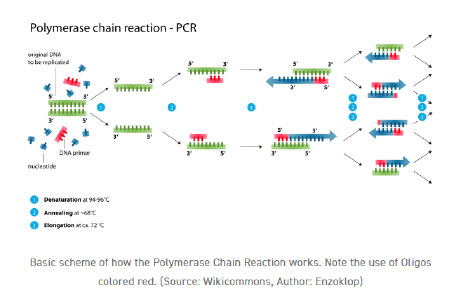
9.2. Sequencing using Oligonucleotides
All modern sequencing systems rely on the same fundamental principle of PCR, which involves the binding of a primer to single-stranded DNA followed by extension, but they all use various technologies to read the resultant sequence of bases. When general primers will do, we choose those over those designed for a specific target. Cloned DNA can be sequenced by using universal primers whose sequences are complementary to those surrounding the multiple cloning site (MCS) of the carrier plasmid. Sequencing is performed using a universal primer and oligonucleotides that have been ligated to the ends of each nucleic acid fragment.14
9.3. Use of Oligonucleotides in Gene Expression Studies
The usage of oligonucleotides as molecular barcode probes is an emerging use of these molecules. These probes use molecular barcode technology, such as that used in NanoString's nCounter® Pro Analytic System, to identify and quantify target transcripts without the need for enzymatic steps like those used in polymerase chain reaction (PCR). To provide very selective annealing in various sample types and subsequent characterization, a sophisticated hybridization process was instead created.
DNA and RNA oligos are employed in the multipart probes that nCounter Pro employs, making them ideal for gene expression research. Two distinct 50mer DNA oligos are produced, each based on a different 100 bp region of the target gene sequence. To make a backbone that is complementary to a fluorescently tagged RNA molecule, one of the 50mer oligos is ligated to another DNA oligo. To produce a code for each gene of interest, the RNA oligo is fluorescently tagged in a distinct linear order of colours (Geiss). This DNA-RNA hybrid is one half of a probe pair, called the reporter, while the other half, called the capture probe, is made up of a second gene-specific 50mer oligo linked to 3' repeats (Geiss). Target RNA annealed to the reporter and capture probes forms a tripartite structure that can be detected and quantified using the reporter probe's fluorescence when the correct hybridization technique has been carried out.
Because of how reliably oligonucleotide hybridization works, probes can tolerate a wide range of sample types, less-than-ideal RNA quality, and differences in copy number. The sophisticated fluorescent molecular barcoded probes used by nCounter Pro highlight the vital function of oligonucleotides in contemporary biotechnology.15
9.4. Antisense Oligonucleotides
Inhibiting mRNA processing or translation using antisense oligonucleotides is a common method for decreasing protein production. Many ongoing clinical trials use this strategy, including treatments for various forms of cancer. Current indications for antisense treatment include CMV retinitis and familial hypercholesterolemia.
A brief stretch of double-stranded DNA is formed when an antisense oligonucleotide's sequence is complementary to a segment of a target messenger RNA. Unmodified or modified antisense oligonucleotides exist. The mechanism(s) of suppressing protein synthesis, the oligonucleotides' ability to penetrate cells, and their susceptibility to degradation are all affected by whether or not the oligonucleotide has been changed.
Mechanisms of inhibition include:
- Degradation of the mRNA double helix by RNase H (unmodified)
- Double-stranded section stalls the translation machinery (all)
- Preventing ribosomal initiation complex binding or RNA splicing protein activity (morpholinos)
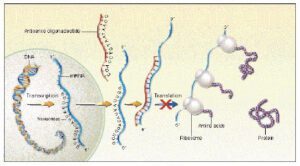
Antisense Oligonucleotides targeted therapy
The most effective morpholino oligonucleotides for therapeutic antisense applications at the present time are those that target sequence upstream of the AUG translation start point or within 30 base pairs downstream of the AUG.16
9.5. Oligonucleotide Therapeutics
Chemically produced oligonucleotides having a single-stranded deoxyribonucleic acid (DNA) or ribonucleic acid (RNA) backbone are used in oligonucleotide therapies, a broad term encompassing cutting-edge molecular-target medicines. By binding to a particular sequence in a target gene or protein, these drugs can suppress the expression of that gene or impair that protein's function.17 As a result, novel treatments against malignancies and genetic illnesses have been developed thanks to the remarkable specificity of oligonucleotide therapy in targeting molecules like mRNA and noncoding RNA that cannot be regulated by traditional pharmaceuticals. In terms of oligonucleotide therapies, antisense oligonucleotides (ASOs), small interfering RNA (siRNA), microRNAs (miRNAs), aptamers, and decoys are all good examples. ASOs and siRNAs, in particular, have advanced further than the others.
Because of its high refractoriness to any existing therapeutic techniques and the difficulty in detecting it in its early stages, pancreatic cancer has been widely acknowledged as a malignant tumour with a bad prognosis in recent review papers.18
With no effective therapies for long-term survival and an upward trend in illnesses, pancreatic cancer is attracting significant focus in pharmaceutical agent development. Because of this, oligonucleotide therapeutic applications for people with pancreatic cancer have already begun in the clinic. Patients with pancreatic cancer have participated in nine NIH-approved clinical studies using oligonucleotide therapies; no such trials have used aptamers or decoys. The National Institutes of Health have given the go-light to a Chinese clinical trial using miRNA-25 for the detection of pancreatic cancer. Patients with pancreatic cancer showed encouraging results in a Japanese clinical study using carbohydrate sulfotransferase 15 (CHST15)-siRNA (STNM01). Previous work of ours has shown that the enzyme CHST15, which biosynthesizes chondroitin sulfate-E (CS-E), is a critical factor in tumor growth by inhibiting tumor cell proliferation in vitro and tumor growth in vivo.19
9.6. Unraveling cellular function with oligonucleotides is a key step toward creating new drugs.
Normal gene function has been studied by studying what happens to a cell or organism when a gene is altered or removed. If a gene responsible for a cell's usual square form is missing, for some reason the cell takes on a circular shape instead. What if, however, the scientist has no notion where in the genome the relevant genes are located or which genes are responsible for a certain function? I was wondering what criteria they use to determine which genes to eliminate. In this context, massively parallel oligo synthesis becomes a crucial tool.
Gene elimination requires the use of genome-editing technologies. The most effective tool for the job is a technique called CRISPR. The CRISPR system consists of a protein that serves as "nano-scissors," snipping DNA like a ribbon and a nucleotide-based guide that informs the scissors exactly where to cut. Usually, the length of a guide is around 100 bases, making it possible for a single oligo to encode the whole guide. With this method, scientists may surgically remove specific DNA sequences from cells.
With the use of modern methods of DNA synthesis, it is possible to generate a large pool of oligonucleotides that, when transcribed, will provide enough splicing instructions to completely eradicate all of the genes in a cell's genome. So, scientists may delete as many genes as they wish, one by one, until they find the one(s) responsible for the behavior of interest. To identify the genes responsible for a certain biological process, millions of cells are equipped with CRISPR scissors and a personalized map drawn from this massive database. When activated, CRISPR generates a cell "library" in which individual cells have had specific genes removed from their DNA. Cells exhibiting aberrant behavior for the targeted function are then identified by researchers. Then, the genomic sequence of the aberrant cell is evaluated to determine which genes were removed during CRISPR editing.
Using this strategy, researchers are uncovering the mutated genes at the root of deadly illnesses like cancer and Alzheimer's for which there are currently no effective treatments. Then, the defective genes are introduced into a cell culture in the lab to create a replica of the illness. By identifying the elements of the model cell that aren't working as they should, scientists may work towards the goal of creating innovative treatments that could one day put an end to many dreadful illnesses.20
9.7. World’s data can be stored in oligonucleotides for a 10,000 years
The most cutting-edge usage of oligonucleotides, which has nothing to do with living things, is to store digital information.
Nowadays, digital data is stored on massive banks of magnetic tape, and is retrieved slowly by robots, much like a large jukebox. When it comes to archiving information, this method is not a good option. As the amount of digital data being created increases, this method will become unsustainable due to the high resource and space costs involved. The cassettes also only last for around a decade. It's imperative that a fresh duplicate of the whole archive be created on a regular basis, notwithstanding the enormous cost involved.
Digital information may now be stored in DNA sequences thanks to a joint effort by Microsoft, Twist Bioscience, and the University of Washington. Data can be encoded into quaternary data and kept in DNA as A, T, C, and G nucleotides as an alternative to the binary data (ones and zeros) often found on cassettes.
DNA is an astonishingly useful data storage medium. One gramme of DNA has a storage capacity of around one zettabyte. To be precise, that's one trillion billion bytes, or 1021 bytes. DNA oligos are durable for thousands of years, take up relatively little space, and are a promising contender for long-term data storage because of these qualities.
The oligos would be processed through a DNA sequencer in order to retrieve the information contained in this fashion. Once you know the sequence, you can use a decoder to get the original binary data. Although the methods for both encoding (by writing millions of oligos) and decoding (by sequencing DNA) already exist, it is up to researchers to refine and combine them into a practical and cost-effective method of storing data. Further promising developments in this area can be anticipated. If you're interested in reading more about this, we covered it in depth in a recent blog article.21
9.8. Other uses of Oligonucleotides
Oligonucleotides have several applications in the fields of science and medicine. Common uses include aptamer creation, allele-specific testing, and dsDNA binding using triplex-forming oligonucleotides.
- Aptameric Nucleic Acids: DNA or RNA aptamers are sequences having a high affinity and specificity for binding to their target. The tertiary structure (helices, etc.) is responsible for the specificity, rather than the sequence itself. Systematic evolution of ligands by exponential enrichment is used to select aptamers by several rounds of enrichment of randomly chosen oligonucleotides or library fragments (SELEX). Stability can be increased by conjugating aptamers with nanoparticles or medicines.
Sensors, regulators of biological processes, and therapeutic tools are all applications for aptamers. Protein function can be inhibited by aptamers due to their high-affinity binding. Drug delivery via aptamers is possible because they attach to proteins found on cell surfaces. Many aptamers, mostly RNA aptamers, are now undergoing human clinical studies for a variety of illnesses.
- Allele-specific oligonucleotides (ASOs): Probes made from allele-specific oligonucleotides (ASOs) are commonly employed in dot-blots to identify variant alleles in DNA. The oligonucleotides are typically between 15 and 20 nucleotides in length and are employed mostly in diagnostics for illnesses caused by frequent point mutations such cystic fibrosis and sickle cell anemia. Mutation detection is achieved by comparing the binding of two oligonucleotides, one with the wild-type sequence and the other with the mutation.
- Triplex-Forming Oligonucleotides (TFOs): Oligonucleotides that form triplexes in DNA, are designed to bind specifically to the main groove. They attach to lengthy sequences of purines and pyrimidines in DNA and typically range in length from 10 to 30 nucleotides. Transcriptional repressors (TFOs) are being studied for their potential application in gene modification by targeted recombination to generate stable, heritable alterations to DNA.22
10. What’s next?
Without the oligonucleotide, where would modern science be? Without oligonucleotides, it would be very challenging to develop novel medications past the first stages, as they allow us to create new cell lines for testing therapies. It may be argued that our knowledge of human ailments, many of which we can now effectively cure, would be severely lacking. Silicon-based DNA synthesis technology, such as that developed by Twist Bioscience, has made it feasible to mass-produce high-quality oligos in unprecedented quantities. The short answer to "what is an oligo?" is that it is a DNA molecule that has changed the world and has a promising future.
References:
- https://www.news-medical.net/life-sciences/What-is-an-Oligonucleotide.aspx
- Monga I, Qureshi A, Thakur N, Gupta AK, Kumar M (2017). "ASPsiRNA: A Resource of ASP-siRNAs Having Therapeutic Potential for Human Genetic Disorders and Algorithm for Prediction of Their Inhibitory Efficacy". G3: Genes, Genomes, Genetics. 7 (9): 2931–2943.
- Weiss B, Davidkova G, Zhou LW (1999). "Antisense RNA gene therapy for studying and modulating biological processes". Cellular and Molecular Life Sciences. 55(3): 334–58.
- https://www.mt.com/gb/en/home/applications/L1_AutoChem_Applications/L2_ReactionAnalysis/oligonucleotide-synthesis.html
- https://nanostring.com/blog/what-is-an-example-of-an-oligonucleotide/
- https://www.aatbio.com/resources/faq-frequently-asked-questions/How-are-oligos-made
- Beaucage, S. L.; Iyer, R. P. (1992). "Advances in the Synthesis of Oligonucleotides by the Phosphoramidite Approach". Tetrahedron. 48 (12): 2223.
- https://www.bio-rad.com/en-uk/applications-technologies/oligonucleotides-design-applications?ID=MOYTCO15
- https://byjus.com/neet/oligosaccharides-examples/#:~:text=Three%20examples%20of%20oligosaccharides%20are,Galactooligosaccharides%20are%20dairy%2Dderived%20oligosaccharides
- https://nanostring.com/blog/what-is-an-example-of-an-oligonucleotide/
- https://www.bio-rad.com/en-uk/applications-technologies/oligonucleotides-design-applications?ID=MOYTCO15
- https://www.biosyn.com/faq/What-are-Oligonucleotides.aspx
- https://www.mt.com/gb/en/home/applications/L1_AutoChem_Applications/L2_ReactionAnalysis/oligonucleotide-synthesis.html
- https://www.bio-rad.com/en-uk/applications-technologies/oligonucleotides-design-applications?ID=MOYTCO15#:~:text=In%20addition%20to%20their%20use,and%20even%20as%20drug%20carriers.
- https://www.biosyn.com/faq/What-are-Oligonucleotides.aspx
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L, Dimitrov K. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008 Mar;26(3):317-25.
- Scherman D., Rousseau A., Bigey P., Escriou V. Genetic pharmacology: Progresses in siRNA delivery and therapeutic applications. Gene Ther. 2017;24:151. doi: 10.1038/gt.2017.6.
- McGuigan A., Kelly P., Turkington R.C., Jones C., Coleman H.G., McCain R.S. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. WJG. 2018;24:4846–4861. doi: 10.3748/wjg.v24.i43.4846.
- Takakura K., Shibazaki Y., Yoneyama H., Fujii M., Hashiguchi T., Ito Z., Kajihara M., Misawa T., Homma S., Ohkusa T., et al. Inhibition of Cell Proliferation and Growth of Pancreatic Cancer by Silencing of Carbohydrate Sulfotransferase 15 In Vitro and in a Xenograft Model. PLoS ONE. 2015;10:e0142981. doi: 10.1371/journal.pone.0142981.
- https://www.twistbioscience.com/blog/perspectives/how-oligos-changed-world#:~:text=Oligos%20are%20short%2C%20synthetic%20strands,such%20as%20those%20in%20DNA.
- https://www.twistbioscience.com/blog/perspectives/how-oligos-changed-world#:~:text=Oligos%20are%20short%2C%20synthetic%20strands,such%20as%20those%20in%20DNA.
- https://www.bio-rad.com/en-uk/applications-technologies/oligonucleotides-design-applications?ID=MOYTCO15#:~:text=In%20addition%20to%20their%20use,and%20even%20as%20drug%20carriers.
